Introduction
In the data science field, databases with large amounts of variables are routinely encountered. In most cases, the size of the database can be reduced by removing highly correlated or superfluous variables. The accuracy and reliability of a classification or prediction model produced from this resultant database will be improved by the removal of these redundant variables. In addition, superfluous variables increase the data-collection and data-processing costs of deploying a model on a large database. One of the first steps in data science is finding a way to reduce the number of independent or input variables used in the model (known as Dimensionality Reduction) without sacrificing accuracy.
Dimensionality Reduction is the process of reducing the amount of variables to be used as input in a prediction or classification model. This domain is divided into two branches: feature selection and feature extraction. Feature selection attempts to discover a subset of the original variables, while feature extraction attempts to map a high-dimensional model to a lower-dimensional space.
Principal Component Analysis (PCA) is a mathematical procedure that transforms a number of (possibly) correlated variables into a smaller number of uncorrelated variables called principal components. The objective of principal component analysis is to reduce the dimensionality (number of variables) of the data set, but retain as much of the original variability in the data as possible. The first principal component accounts for the majority of the variability in the data, the second principal component accounts for the majority of the remaining variability, and so on.
A PCA is concerned with explaining the variance covariance structure of a high dimensional random vector through a few linear combinations of the original component variables. Consider a database X with m rows and n columns (4 x 3)
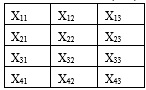
The first step in reducing the number of columns (variables) in the X matrix using the PCA algorithm is to find the mean of each column.
Next, the algorithm subtracts each element in the database by the mean (Mu), thereby obtaining a new matrix X that contains 4 rows and 3 columns.

The PCA algorithm calculates the covariance or correlation matrix (depending on the user's preference) of the new X matrix.
The algorithm calculates eigenvalues and eigenvectors from the covariance matrix for each variable and lists these eigenvalues in order from largest to smallest. Larger eigenvalues denote that the variable should remain in the database. Variables with smaller eigenvalues will be removed according to preference.
Analytic Solver Data Science chooses between selecting a fixed number of components (variables) to be included in the 'reduced' matrix (referred to as Y matrix) or the smallest subset of variables that explains or accounts for a certain percentage variance in the database. Variables with eigenvalues below the chosen threshold will not be included in the Y matrix. Assume that a fixed number of variables (2) be included in the Y matrix.
A new matrix V is formed, containing eigenvectors based on the selected eigenvalues.
The original matrix X that has 4 rows and 3 columns will be multiplied by the V matrix, containing 4 rows and 2 columns. This matrix multiplication results in the new reduced Y matrix, containing 4 rows and 2 columns.
In algebraic form, consider a p-dimensional random vector X = ( X1, X2, ..., Xp ) where p principal components of X are k univariate random variables Y1, Y2, ..., Yk, defined by the following formula:

where, the coefficient vectors l1, l2 ,..etc. are chosen such that they satisfy the following conditions:
First Principal Component = Linear combination l1'X that maximizes Var(l1'X) and || l1 || =1
Second Principal Component = Linear combination l2'X that maximizes Var(l2'X) and || l2 || =1
and Cov(l1'X , l2'X) =0
jth Principal Component = Linear combination lj'X that maximizes Var(lj'X) and || lj || =1
and Cov(lk'X , lj'X) =0 for all k < j
These functions indicate that the principal components are those linear combinations of the original variables that maximize the variance of the linear combination and have zero covariance (i.e., zero correlation) with the previous principal components.
It can be proved that there are exactly p such linear combinations. However, typically, the first few principal components explain most of the variance in the original data. As a result, instead of working with all the original variables X1, X2, ..., Xp, first perform PCA, and then use only the first two or three principal components in a subsequent analysis.
